Speaker: Jérôme Feret ENS Ulm, jerome.feret@ens.psl.eu
Tuesday Mar 26 2024, 14:00, Room 1Z56
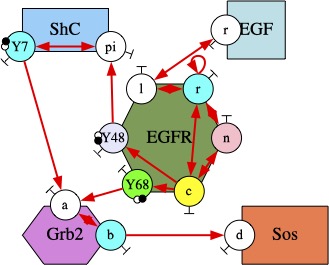
Abstract: Software sciences have a role to play in the description, the organization, the execution, and the analysis of the molecular interaction systems such as biological signaling pathways. These systems involve a huge diversity of bio-molecular entities whereas their dynamics may be driven by races for shared resources, interactions at different time- and concentration-scales, and non-linear feedback loops. Understanding how the behavior of the populations of proteins orchestrates itself from their individual interactions, which is the holy grail on systems biology, requires dedicated languages offering adapted levels of abstraction and efficient analysis tools.
In this talk we describe the design of formal tools for Kappa, a site-graph rewriting language inspired by bio-chemistry. In particular, we introduce a static analysis to compute some properties on the biological entities that may arise in models, so as to increase our confidence in them. We also present a model reduction approach based on a study of the flow of information between the different regions of the biological entities and the potential symmetries. This approach is applied both in the differential and in the stochastic semantics.



